In 2020, Imec submitted a paper and it was published in Sensors (Sensors 2020, 20(24), 7302). The paper was published as part of the “ITEA Inno4Health 19008 project”. See more information about the paper below.
Platform for Analysis and Labeling of Medical Time Series
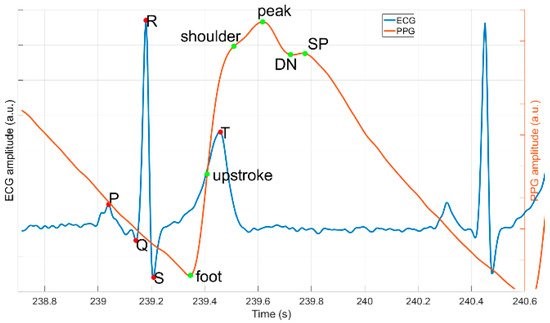
Abstract: Reliable and diverse labeled reference data are essential for the development of high-quality processing algorithms for medical signals, such as electrocardiogram (ECG) and photoplethysmogram (PPG). Here, we present the Platform for Analysis and Labeling of Medical time Series (PALMS) designed in Python. Its graphical user interface (GUI) facilitates three main types of manual annotations—(1) fiducials, e.g., R-peaks of ECG; (2) events with an adjustable duration, e.g., arrhythmic episodes; and (3) signal quality, e.g., data parts corrupted by motion artifacts. All annotations can be attributed to the same signal simultaneously in an ergonomic and user-friendly manner. Configuration for different data and annotation types is straightforward and flexible in order to use a wide range of data sources and to address many different use cases. Above all, configuration of PALMS allows plugging-in existing algorithms to display outcomes of automated processing, such as automatic R-peak detection, and to manually correct them where needed. This enables fast annotation and can be used to further improve algorithms. The GUI is currently complemented by ECG and PPG algorithms that detect characteristic points with high accuracy. The ECG algorithm reached 99% on the MIT/BIH arrhythmia database. The PPG algorithm was validated on two public databases with an F1-score above 98%. The GUI and optional algorithms result in an advanced software tool that allows the creation of diverse reference sets for existing datasets.
Read the full paper via: https://www.mdpi.com/1424-8220/20/24/7302.